Experimental strategies for null allele generation
A proven strategy for understanding gene functions is to remove the gene or its protein product and perform a set of assays to study how its loss affects molecular and cellular phenotypes. The Data Production Centers have employed the following strategies for null allele generation:
1. Gene knock-out (KO)
KO - Premature termination codon (PTC)
Strategy used by: The Jackson Laboratory, view on protocols.io
The PTC+1 strategy involves precise CRISPR-Cas9 engineering of a premature stop codon and the insertion of a degenerate base in an early common exon, thereby truncating all isoforms of the protein and introducing a frame-shift mutation to ensure the production of a non-functional protein, essentially "knocking out" the normal function of the gene. Depending on the position of the PTC+1 mutation with respect to the genomic structure of the gene, the mutant mRNA may or may not be subject to nonsense mediated decay (NMD).

Strategy used by: Memorial Sloan Kettering Cancer Center
Frameshift indel mutations introduced by CRISPR-Cas9 typically result in the creation of a premature stop codon within the coding region, producing a truncated, nonfunctional protein and effectively "knocking out" the gene’s normal function. At MSK, they have generated knockout hPSC lines, and a small number of enhancer deletion lines. Then they individually barcoded these lines, and pooled them for differentiation experiments. Cells were collected at consecutive differentiation stages for scRNA-seq, followed by computational demultiplexing to analyze the transcriptomic phenotypes associated with each individual knockout condition.

KO - Critical exon deletion
Strategy used by: The Jackson Laboratory, view on protocols.io
A critical exon deletion refers to a genetic knockout (KO) where an early common, frame-shifting exon within a gene has been deleted, leading to a truncated, nonfunctional protein being produced; essentially "knocking out" the normal function of the gene. Depending on the position of the premature termination codon with respect to the genomic structure of the gene, the mutant mRNA may or may not be subject to nonsense mediated decay (NMD).

KO - Gene deletion
Strategy used by: The Jackson Laboratory, view on protocols.io
Gene deletion means the complete removal of most or all protein coding exons of the gene, eliminating the possibility of the protein production entirely. The non-coding RNA transcript that remains is not subject to nonsense mediated decay and can potentially be identified by RNA-seq.

KO - Insertion of a cassette (a KI-KO approach)
Strategy used by: Memorial Sloan Kettering Cancer Center
A cassette is knocked into the target gene coding region. This insertion is expected to disrupt transcription due to the large insertion size, introduce frameshift mutations, and create a premature termination codon, thus effectively "knocking out" the normal function of the gene.

2. Protein degradation
Auxin-inducible degron
Strategy used by: Northwestern University
Auxin-inducible degron (AID) is a chemical genetic tool that uses the plant hormone auxin to degrade specific proteins in mammalian cells, which allows researchers to study protein function in living tissues.
At NWU, cells are engineered to express the TIR1 auxin receptor and Crispr-Cas9 technology is used to add the AID protein degron tag to specific endogenous gene loci. Cells express the tagged protein normally until treated with Auxin, which induces rapid, reversible protein degradation.

3. Gene knockdown
CRISPR interference (CRISPRi)
Strategy used by: University of California San Francisco
CRISPRi (CRISPR interference) utilizes a catalytically dead Cas9 (dCas9) protein fused to a KRAB (Krüppel-associated box) domain, a transcriptional repressor. A guide RNA (gRNA) directs dCas9-KRAB to a specific DNA sequence, where it binds and blocks transcription by recruiting endogenous chromatin-modifying repressive complexes via the KRAB domain. This approach enables reversible, precise, and non-destructive repression of gene expression at the transcriptional level, making it ideal for functional genomics and studying essential genes.
DAVs Analysis Methods
Bulk RNAseq Differential Expression Analysis
Strategy used by: Fred Hutch Cancer Center, view on GitHub
To quantify KO effect using Bulk RNA-seq data, we mainly focus on Differential Expression (DE) analysis comparing knock out (KO) versus wild type (WT) samples, using method DESeq2, followed by gene set enrichment analysis.
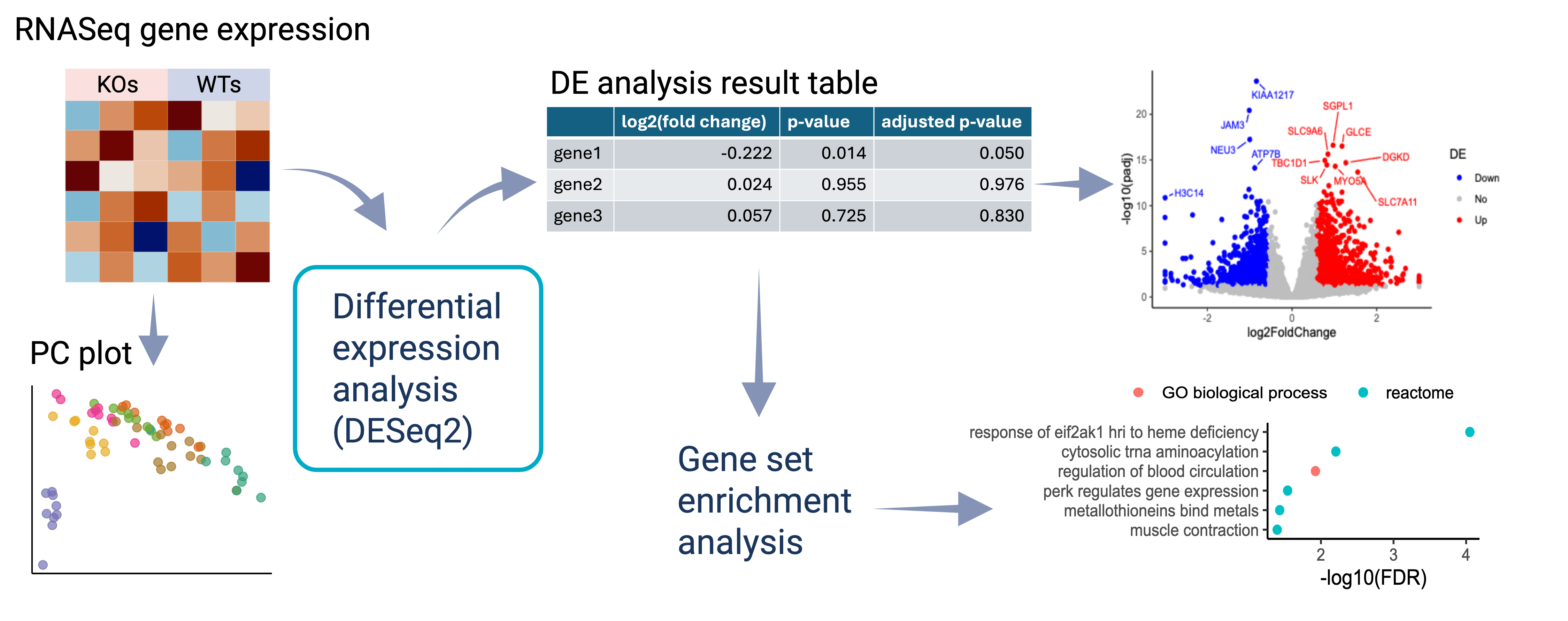
The Jackson Laboratory produced bulk RNAseq data with gene knockout and WT involving different gene knockout strategies, different model systems, different cell lines, and different days. For certain gene, the datasets involve two oxygen conditions.
Each dataset is processed separately. To assess the differential expression (DE) effects of gene knockout, the steps taken include quality control, DE analysis, and functional category enrichment analysis. For data visualization, volcano plots are provided to highlight the significantly differentially expressed genes under each gene knockout, and bar plots are provided to show significantly enriched functional categories. Detailed DE results are provided in tables, including basic information for each gene and the corresponding log2(fold change), p-value, and adjusted p-value.
The gene differential expression analysis method used is DESeq2. The major type of DE testing is between knockout samples and wild type samples. The second type of DE testing is between the two oxygen conditions. Proper levels of sample groups are chosen to run DESeq2 on, balancing between borrowing information across samples and avoiding batch effect.
When running DESeq2 model, there are two covariates considered: sequencing run ID and read depth (computed as the 75% quantile of gene expression in each sample). The sequencing run ID is included if the samples involve more than one sequencing run. To decide whether to include read depth, a model including read depth is fit first, and the proportion of genes for which the read depth can explain a significant amount of variation is estimated. The read depth is included in the final model if the estimated proportion is above a threshold that is adjusted according to sample size.
To make the results more reliable, multiple filtering steps are carried out based on the expression level of the genes. Genes with low expression are excluded from DE testing or have adjusted p-value marked as NA.
scRNA-seq Cell Type Composition
Strategy used by: Fred Hutch Cancer Center, view on GitHub
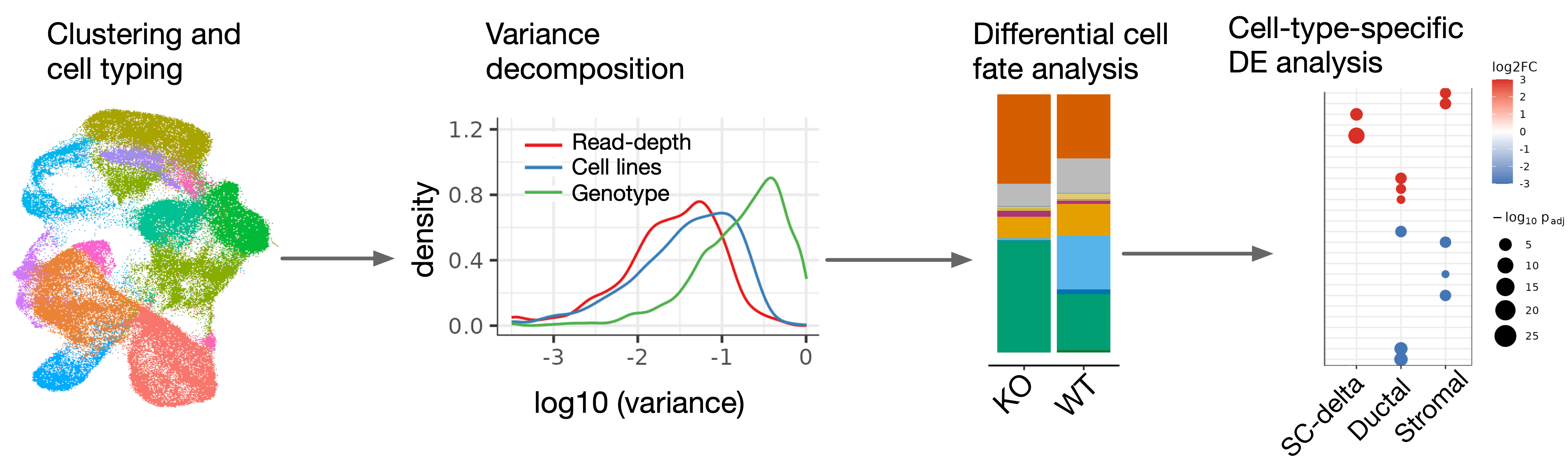
To quantify perturbation effects on cell type composition, pseudobulk samples were generated for each clone at each time point. A linear regression model was then used to model log-transformed cell type proportions, with cell background, data source, and genotype included as covariates.
scRNA-seq Variance Decomposition
Strategy used by: Fred Hutch Cancer Center, view on GitHub
We quantified the proportion of variance explained by read depth, cell background (cell line), and genotype using a series of linear models fitted separately at each time point. For each time point, the models were applied to pseudobulk samples, with log-normalized expression of highly variable genes as the outcome. The genotype effect captures both its influence on cell type composition and on cell type-specific gene expression.
scRNA-seq Differential Expression Analysis - Pseudobulk-based approach
Strategy used by: Fred Hutch Cancer Center, view on GitHub
To identify differentially expressed genes (DEGs) between a KO genotype of interest and WT, pseudobulk samples were generated for each clone–cell type combination at each time point. Differential expression analysis was performed using the DESeq2 package in R, with read depth and cell background as covariates.
scRNA-seq Differential Expression Analysis - Cell-level approach
Strategy used by: Fred Hutch Cancer Center, view on GitHub
Alternatively, for each time point and cell type, cell-level differential expression analysis was performed using FindMarkers from the Seurat R package with default parameters.
Gene program evaluation
Strategy used by: Stanford, view on GitHub
cNMF is a computational method that uncovers groups of genes called “gene programs” that work together inside cells. By repeating the analysis many times and focusing on the most consistent results, it filters out noise and highlights the most meaningful patterns. These gene programs are then tested against biological knowledge to see if they reflect known pathways, cell types, or cellular activities. This combination of discovery and evaluation helps researchers understand what makes cells unique, what activities they are carrying out, and how these processes vary across conditions, development, or disease.
cNMF is a consensus-based matrix factorization approach that decomposes single-cell RNA-seq data into interpretable gene programs. It applies NMF repeatedly with different random initializations, clusters the resulting factors, and aggregates them into consensus programs. This reduces stochastic variability and increases reproducibility compared to single NMF runs. The output consists of a program usage matrix (cells × programs), which describes the activity of each program in each cell, and a gene loading matrix (programs × genes), which identifies the genes that define each program.
To ensure both robustness and biological interpretability, cNMF results are systematically evaluated using multiple criteria:
- Reconstruction error and stability, measuring how well the decomposition explains the data and how reproducible the programs are.
- Co-regulation and coherence, testing whether program genes are functionally linked.
- Biological enrichment, assessing whether programs correspond to known pathways, processes, or cell identities.
- Cross-dataset reproducibility, ensuring that programs are consistent across experimental replicates and biological contexts.
- Through this strategy, cNMF reliably identifies both identity programs (cell-type signatures) and activity programs (dynamic patterns such as cell cycle, stress responses, or signaling).
ChromBPNet
Strategy used by: Stanford, view on GitHub
ChromBPNet is a machine learning model that helps scientists read the “regulatory code” written in our DNA. It learns how DNA sequence shapes the way our genome is packaged and accessed inside cells, while carefully separating out technical artifacts from true biological signals. By doing so, ChromBPNet can pinpoint the short DNA patterns that control when and where genes turn on, and show how these patterns change across cell types and conditions. The model can also predict how small changes in DNA—such as genetic variants—might disrupt gene regulation, offering clues to the biological differences between individuals and insights into the genetic basis of health and disease.
ChromBPNet is a base-resolution convolutional neural network built to decode the cis-regulatory code and predict how DNA sequence controls chromatin accessibility. Its bias-factorized architecture explicitly separates enzyme-specific cleavage bias from true regulatory signal: a “bias model” first learns the sequence preferences of DNase-I or Tn5, and a “TF model” then trains on bias-corrected profiles to capture sequence features that truly drive accessibility. This separation yields highly interpretable results, enabling recovery of transcription factor motif lexicons, cooperative motif syntax, and single-base footprints. ChromBPNet’s compact design matches or exceeds the performance of much larger models in predicting the impact of genetic variants on accessibility, pioneer factor binding, and reporter activity. By providing fine-grained, cell-type–specific insights, ChromBPNet offers a powerful framework for understanding how genomic regulation is encoded in DNA, how it varies across cell types, assays, sequencing depths, and populations, and how genetic variation may contribute to complex traits and disease.
Uniform and Scalable Data Processing Methods by the DRACC
Analytical pipelines
To facilitate reproducible analyses across diverse data types, graphical and interactive analytical pipelines are developed in the open-source Biodepot platform with a training portal available at https://biodepot.github.io/training/. These analytical pipelines are developed by the DRACC with feedback solicited from members in the MorPhiC consortium. Pipelines currently supported include:
- Bulk RNA-seq. GitHub https://github.com/morphic-bio/Bulk-RNA-seq
- gRNA enrichment. GitHub https://github.com/morphic-bio/gRNA-Enrichment. A pre-configured instance is available at https://gitpod.io/#https://github.com/morphic-bio/gRNA-Enrichment
- Single cell/perturb-seq (GitHub https://github.com/morphic-bio/scRNA-seq) with a novel open-source utility called assignBarcodes, designed for targeted sequencing analysis in single-cell experiments (GitHub https://github.com/morphic-bio/process_features). assignBarcodes efficiently assigns feature barcodes from FASTQ files to a known set of sequence barcodes, serving as a powerful, open-source alternative to proprietary tools.
Due to consent restrictions for the KOLF2 cell line and derivatives that require data mapping to the Y chromosome to be removed, a filtering step has been added to our analytical pipelines for all data generated using these cell lines. Specifically, all reads in the BAM and FASTQ files that align to the Y-chromosome are removed, but the counts tables include the Y chromosome data.
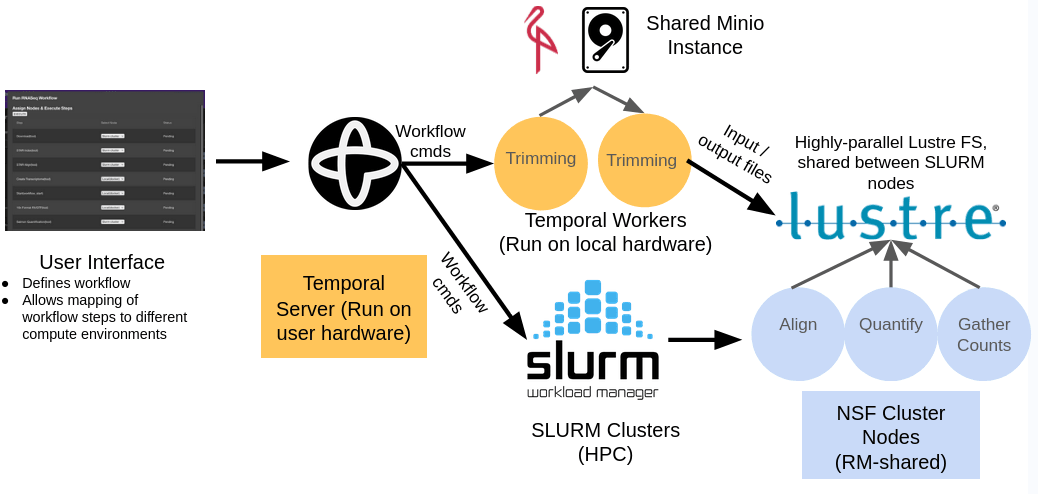
Scalable data processing
A scheduler based on the Temporal.io framework has been developed to enable optimizations of bioinformatics workflows. Specifically, users can transparently map workflow steps to diverse execution environments, including high-performance computing (HPC) resources managed by the SLURM resource manager through an easy-to-use graphical user interface. Asynchronous execution of workflows is supported to optimize resource utilization even when the scheduler cannot make use of a system’s full RAM and CPU resources. Pipelines are executed using a combination of UW compute resources and the NSF Bridges2 supercomputer.
Pre-print: bioRxiv 2025.09.01.673517